As we have seen with Retrieval Augmented Generation (RAG), integrating LLMs with unstructured data can be valuable for a wide range of use cases.
Businesses have gathered years of domain knowledge in silos like Microsoft SharePoint, email, Slack, Microsoft Teams, and Zoom recordings, and they want to explore the hidden relationships between people, organizations, places, and events to provide deeper insights to aid day-to-day decision making.
Knowledge graphs are a perfect fit for modeling these relationships, and LLMs can be used to extract both the entities and their inter-relationships.
In addition, knowledge graphs can be used as context for the RAG pipeline, in addition to content retrieved by 'classic' embeddings-based semantic search
TL;DR
We have published a sample application on GitHub, which shows how we can build a knowledge graph automatically using OpenAI GPT-4o for entity extraction.
You can run the demo yourself here, and log into your SharePoint account to ingest files - documents, audio, images, etc. It requires a free signup on our Developer Portal to get the required keys.
Extracting your Knowledge Graph
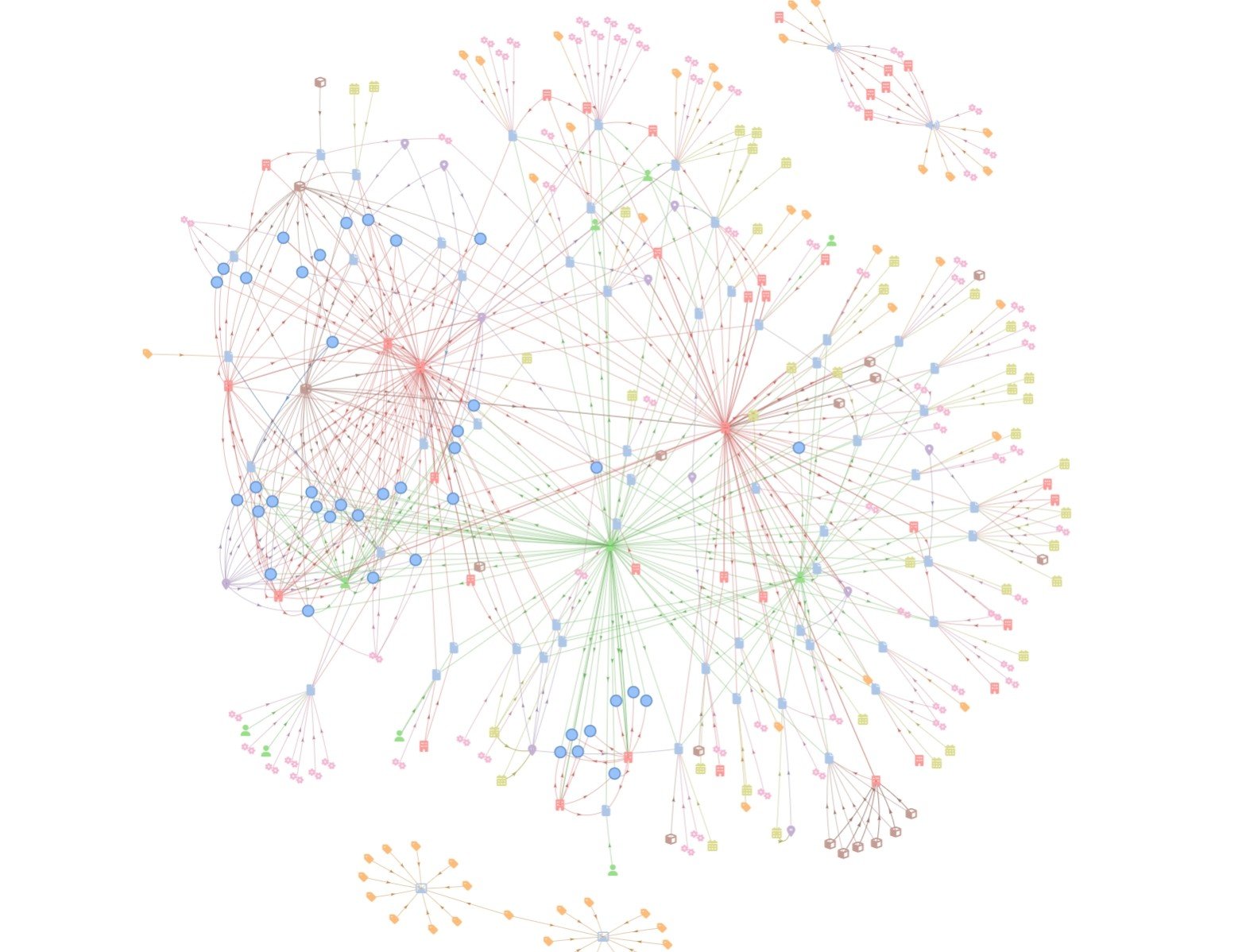
Below is a knowledge graph, with entities automatically extracted from a set of sample data found in a SharePoint folder.
We have used Graphlit to automatically extract images from PDFs, and are using the OpenAI GPT-4 Vision model to perform OCR and generate detailed text descriptions of the images.
Entity extraction is perfomed, not just on textual content, but across multimodal content as well - audio transcripts, image descriptions, etc.
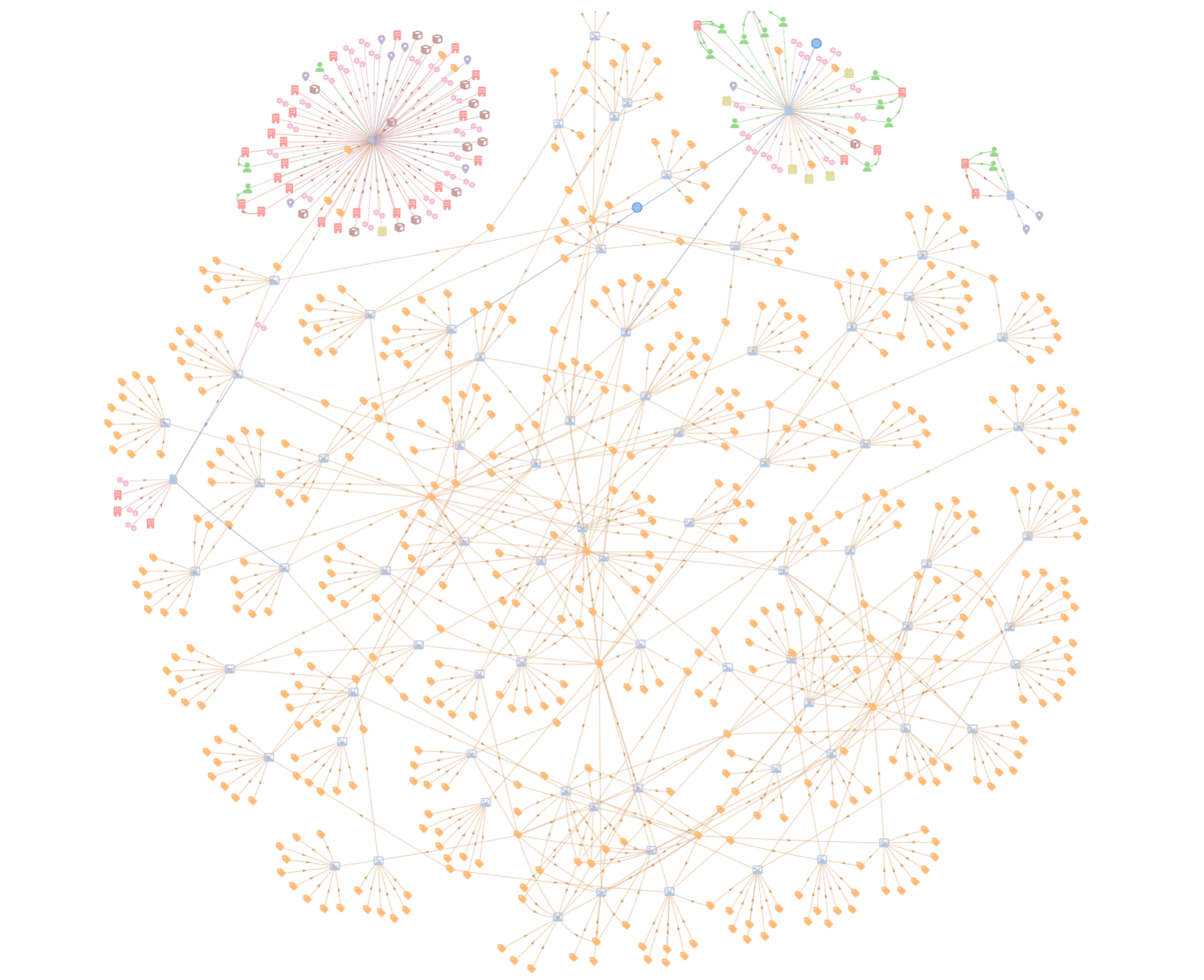
Here we can see the entities extracted from my appearance on the Data Engineering Podcast.
You can see that it has extracted a number of 'organization' entities such as Microsoft, GitHub and Figma, which were discussed on the podcast. These entities were extracted from the audio transcript, which was automatically generated by Graphlit using a Deepgram transcription model.
Most of the entities could be considered 'tags' on the podcast content, and while interesting to visualize, they are valuable to the cataloging of the content - and will be key later for the retrieval element of GraphRAG.
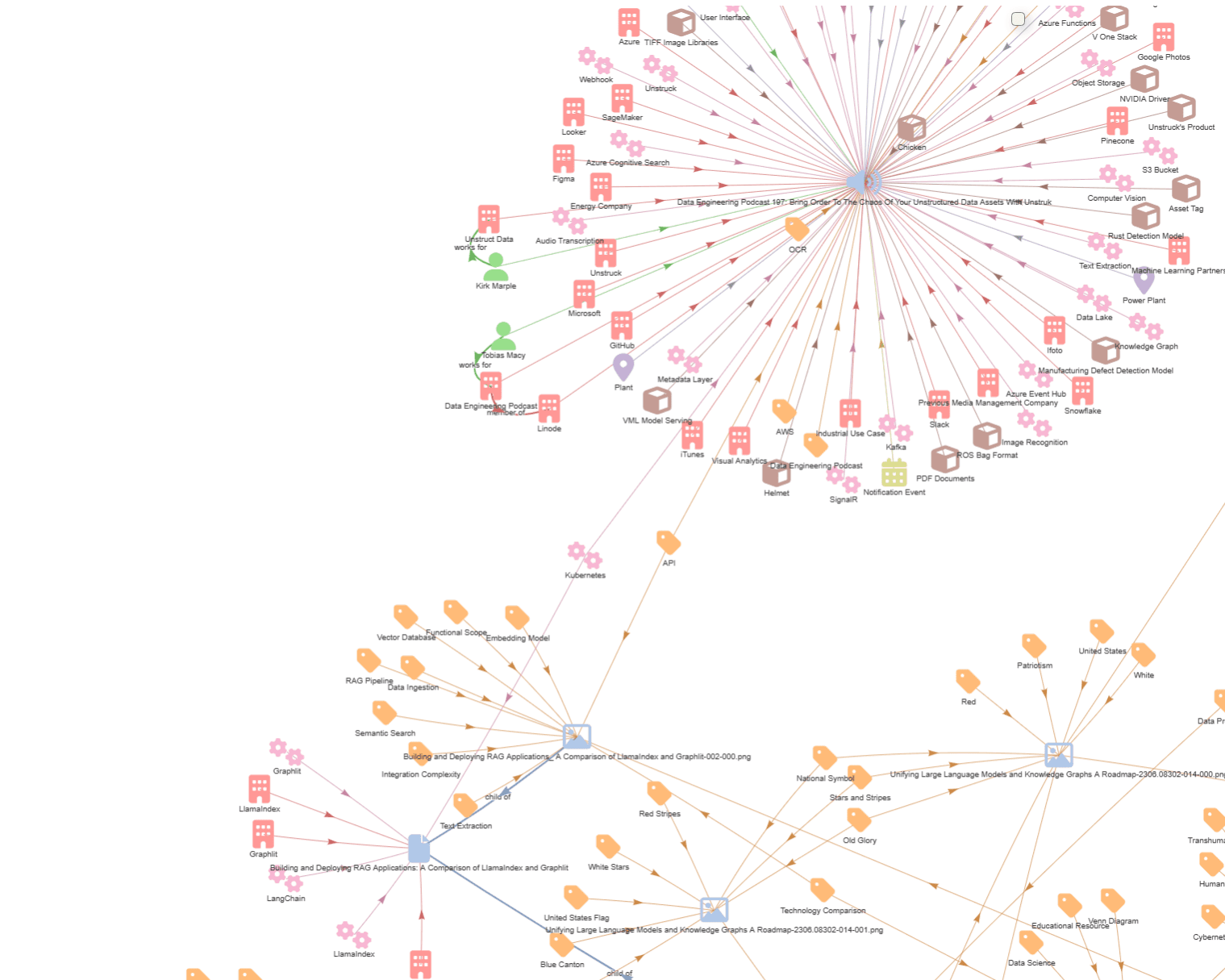
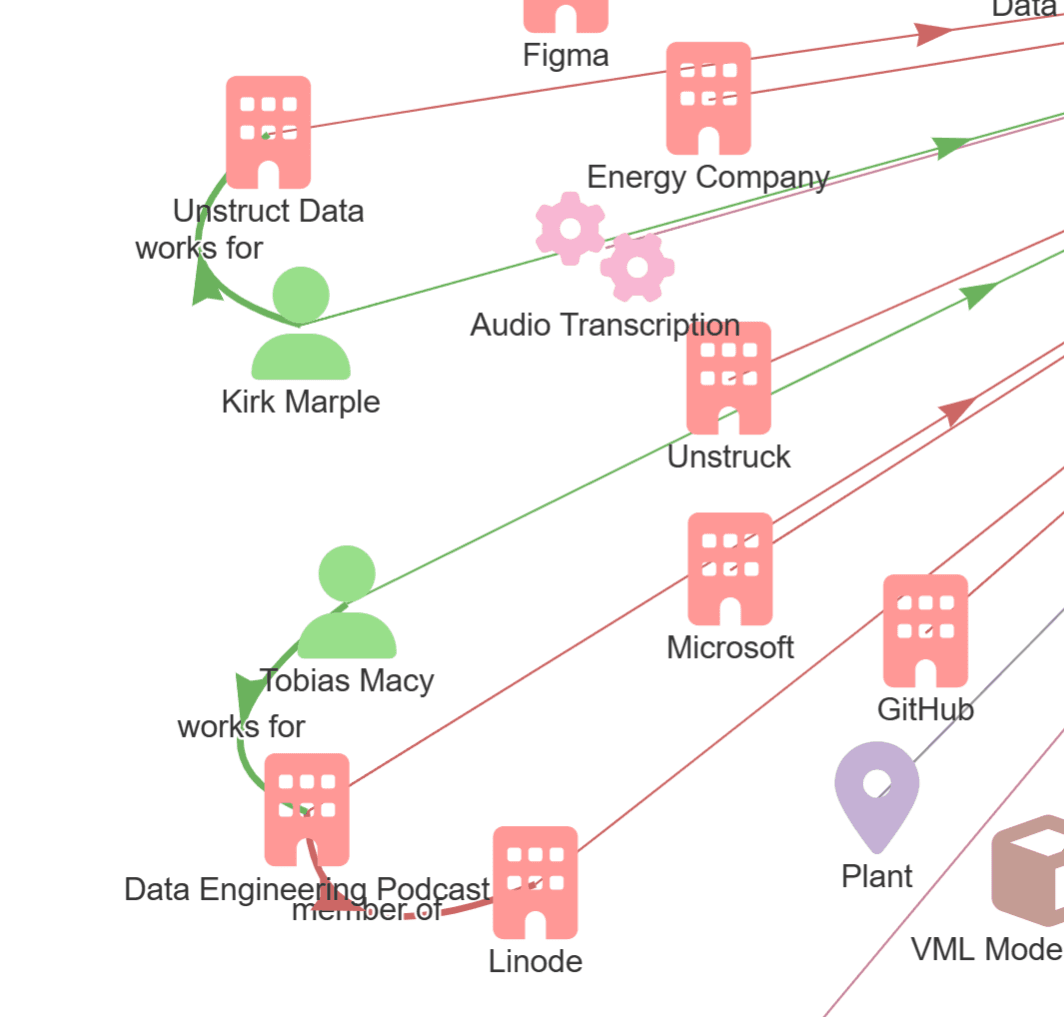
As for focus further in on the relationships, we can see that it found that I (Kirk Marple) work for Unstruct (sic) Data, and Tobias Macy (sic) works for the Data Engineering Podcast, which is a member of Linode. Now, the LLM has identified 'member of' as the relationship, while I believe Linode is actually a sponsor of the podcast.
Extracted entities still need to be reviewed either by humans, or evaluated with other LLMs, possibly fine-tuned on the domain of interest.
Also, entity resolution - or deduplication - is an important step to increase the quality of your knowledge graph relationships by removing duplicate nodes or edges, and by merging similar nodes together (or splitting nodes into separate nodes).
As we've seen, the transcription could have incorrectly named an organization, such as Unstruct Data vs Unstruk Data, and it's important to use similarity and fuzzy searches to identify entities which need to be corrected or merged. We can store aliases in each entity for common misspellings to make identifying these similar entities easier.
GraphRAG vs RAG
When prompting a conversation with an LLM, similar to ChatGPT, the first step is to retrieve content similar to the user's prompt. We can assume that content has been prepared upon ingest, where text is extracted - optimally into semantic chunks - from documents such as PDFs or DOCX, from audio transcripts or even from image descriptions. The extracted text gets chunked - optimally around 500-800 tokens, and overlapped - and has vector embeddings created with a model like OpenAI Ada-002.
Vector databases, such as Azure AI Search, Pinecone or Qdrant, are used for vector-based retrieval using a similarity search algorithm like HNSW. In this method, the vector embeddings are used to return 'k' nearest neighbors around the vector embedding created by the user prompt.
In this way, we get back a ranked list of text chunks, with metadata which links them back to the original content, and which page the text was found on, or what time range it was found in the transcript.
We can also use a hybrid approach of BM25 keyword retrieval with vector retrieval, which can benefit certain retrieval use cases.
The retrieved text is formatted into the final prompt, which is sent to the LLM for completion. This final prompt, we call the 'prompt context'. It can contain previous messages in the current conversation thread, as well as content text chunks formatted with useful metadata. It also will contain instructions and guidance for the LLM so it provides the most accurate and relevant response.
Also, we can instruct the LLM to return a structured data format, like JSON, so we're able to the text of the response in addition to citations or other structured data. This also gives us the ability to provide 'guardrails' where we can guarantee the response matches a desired JSON schema, and has not been truncated.
Most RAG pipelines use a similar approach, as described here, and GraphRAG introduces a new pattern which can enrich the retrieval stage of the pipeline and provide greater 'color' about the referenced entities in the LLM response.
GraphRAG as a Pattern, not a Product
GraphRAG is not a product, per se, but is a pattern which extends the standard RAG pipeline. Other vendors such as Microsoft and Neo4j have published their interpretations of GraphRAG, which are complementary to the Graphlit approach. Graphlit's difference, as a managed platform, is that GraphRAG is configurable within an existing Graphlit LLM specification. It can be enabled by adding a graphStrategy field, which specifies the details of how the graph will be used in the GraphRAG pipeline.
There are two ways that Graphlit leverages the knowledge graph during the RAG pipeline: filtering and context.
Using the Knowledge Graph for Filtering
Once we have the knowledge graph extracted from ingested content, we can use it for filtering content during retrieval.
We take the user prompt, such as "When did Kirk Marple work for Microsoft in Seattle?", and use an LLM to extract named entities. Currently we are using Anthropic Claude 3 Haiku by default for this, because of its quick performance for LLM responses, but any LLM can be used.
With our entity extraction process, it returns three entities from the user prompt: "Kirk Marple", person; "Microsoft", organization; and "Seattle", place.
We automatically assign 'observation filters' for these 'observed entities', where the semantic search will only return content which has observed these three entities. Observations are edges in the knowledge graph between a piece of content, like a PDF, and a named entity, like a Person.
This can be a useful way to trim down a large corpus of ingested content, during retrieval, so the vector search is looking for embedding similarity on a smaller dataset.
Filtering by observations works in conjunction with metadata filtering that is also supported by the standard RAG pipeline.
Once we have the resulting set of filtered context, we compile the prompt context for the LLM, and send it for completion.
Using the Knowledge Graph for Context
In addition to using the knowledge graph for filtering, we can leverage the extracted entities to provide greater context for the LLM response, than what was provided in the extracted text.
We use a process called 'entity enrichment' where external APIs, such as Crunchbase or Wikipedia, can be queried with the entity name to return additional metadata about the entity. For example, we can query Wikipedia about Microsoft, and get back metadata about the number of employees and revenue, in addition to a detailed text description.
When processing the GraphRAG pipeline, after retrieving the similar contents, we can query the entities extracted from the user prompt, and add their metadata to the prompt context.
In this way, we're giving more 'color' to the prompt context about the entities that the user is interested in.
When using LLMs for research, this has been shown to give a higher quality response than the standard RAG pipeline. By providing extra detail that exists in the entity graph, and not in the extracted text, we are able to have the LLM respond with background information and details otherwise not possible with standard RAG.
Using standard RAG:

Using GraphRAG:
Using the Knowledge Graph for Visualization
After prompt completion, we have a set of filtered contents which were used for the RAG pipeline.
These contents may also have observations of extracted entities, such as persons or organizations.
When responding with the LLM completion, we automatically generate a knowledge graph for visualization by the end-user application using Graphlit. By creating graph nodes for the contents, and any observed entities, and by creating graph edges for the observations and inter-entity relationships, we have all the data required for data visualization. We are also storing content and entity metadata with the graph nodes, which can be helpful for visualization (i.e. coloring, formatting) of the nodes.
GraphRAG Demo
We have published a sample application on GitHub, which shows how we can build a knowledge graph automatically using OpenAI GPT-4o for entity extraction.
You can run the demo yourself here, and log into your SharePoint account to ingest files - documents, audio, images, etc. It requires a free signup on our Developer Portal to get the required keys.
Code Walkthrough
Let's walk through the code required to build the demo, and see how to create an entity extraction workflow, visualize a knowledge graph, and create a GraphRAG conversation.
This application uses Streamlit to provide the user interface, but the concept described here can be used in your own web application, using our Python or Node.JS SDKs.
Entity Extraction Workflow
Before ingesting our content from SharePoint, we need to provide instructions to Graphlit on how to prepare and extract the files we will ingest, and enrich the entities extracted from the content.
This is done by creating a content workflow object, and typically these are created once for your application, and shared across feeds.
The content workflow may look complicated, but it's straightforward to understand.
Content workflows are handled in sequential stages: preparation, extraction and enrichment.
Here we are saying to use the LAYOUT model from Azure Document Intelligence for PDF OCR and semantic chunking, during text extraction. For documents, we are saying to extract images and store as child contents. For example, we will extract images from PDFs, and link them to the parent document.
Once content has been prepared, we are saying to extract entities with an LLM, i.e. MODEL_TEXT, and to use OpenAI GPT-4 Vision model to extract text and generate descriptions from images.
Last, we are saying to enrich any extracted entities with Wikipedia, which can provide useful metadata and descriptions for the GraphRAG pipeline.
async def create_workflow():
input = WorkflowInput(
name="Workflow",
preparation=PreparationWorkflowStageInput(
jobs=[
PreparationWorkflowJobInput(
connector=FilePreparationConnectorInput(
type=FilePreparationServiceTypes.AZURE_DOCUMENT_INTELLIGENCE,
azureDocument=AzureDocumentPreparationPropertiesInput(
model=AzureDocumentIntelligenceModels.LAYOUT
)
)
),
PreparationWorkflowJobInput(
connector=FilePreparationConnectorInput(
type=FilePreparationServiceTypes.DOCUMENT,
document=DocumentPreparationPropertiesInput(
includeImages=True
)
)
)
]
),
extraction=ExtractionWorkflowStageInput(
jobs=[
ExtractionWorkflowJobInput(
connector=EntityExtractionConnectorInput(
type=EntityExtractionServiceTypes.MODEL_TEXT
)
),
ExtractionWorkflowJobInput(
connector=EntityExtractionConnectorInput(
type=EntityExtractionServiceTypes.OPEN_AI_IMAGE,
openAIImage=OpenAIImageExtractionPropertiesInput(
detailLevel=OpenAIVisionDetailLevels.HIGH
)
)
)
]
),
enrichment=EnrichmentWorkflowStageInput(
jobs=[
EnrichmentWorkflowJobInput(
connector=EntityEnrichmentConnectorInput(
type=EntityEnrichmentServiceTypes.WIKIPEDIA
)
)
]
)
)
graphlit: Optional[Graphlit] = st.session_state['graphlit']
try:
response = await graphlit.client.create_workflow(input)
st.session_state['workflow_id'] = response.create_workflow.id
except GraphQLClientError as e:
return str(e)
return None
Authentication to Microsoft Graph API
Authenticating to any Microsoft resource, such as SharePoint, Microsoft Teams or Microsoft Email, requires the msal (Microsoft Authentication Library) in Python.
This provides the ability to acquire a token, usable against the Microsoft Graph API, based on the scopes you have specified.
Here we are asking for just the User.Read and Files.Read.All scopes, which are required for enumerating and reading SharePoint document libraries, folders and files.
You will need to register an application with your Microsoft Entra ID resource, and get the client ID and secret for your application. In addition, you will need to add the redirect URI for your application, so Microsoft can redirect back to your application after login. (For local development, you can provide a URI like https://localhost:8051.)
In Streamlit, we accept the redirection by checking the query parameters for code, and then we know it's calling us back with the authentication code, and we can trade that for an access token.
We store the refresh token provided by Microsoft to pass to Graphlit for authenticating against the Microsoft Graph API.
SCOPES = ["User.Read", "Files.Read.All"]
redirect_uri = st.secrets["default"]["redirect_uri"]
if not st.session_state['app']:
app = ConfidentialClientApplication(
st.secrets["default"]["client_id"],
authority=f'https://login.microsoftonline.com/common',
client_credential=st.secrets["default"]["client_secret"]
)
st.session_state['app'] = app
if st.query_params and 'code' in st.query_params:
app = st.session_state['app']
code = st.query_params['code']
if st.session_state.refresh_token is None:
result = app.acquire_token_by_authorization_code(
code,
SCOPES,
redirect_uri=redirect_uri
)
if 'access_token' in result:
st.session_state.refresh_token = result['refresh_token']
else:
st.error("Failed to get access token.")
SharePoint Ingestion
Once we have the refresh token, we can list the SharePoint document libraries available to the authenticated user.
This provides the ability to dynamically select the document library, and folder within the library, within your Streamlit application. Graphlit provides helper functions for enumerating these, which internally talks to the Microsoft Graph API.
async def query_sharepoint_libraries():
graphlit: Optional[Graphlit] = st.session_state['graphlit']
input = SharePointLibrariesInput(
authenticationType=SharePointAuthenticationTypes.USER,
refreshToken=st.session_state['refresh_token']
)
try:
response = await graphlit.client.query_share_point_libraries(input)
return response.share_point_libraries.account_name, response.share_point_libraries.results, None
except GraphQLClientError as e:
return None, None, str(e)
async def query_sharepoint_folders(libraryId):
graphlit: Optional[Graphlit] = st.session_state['graphlit']
input = SharePointFoldersInput(
authenticationType=SharePointAuthenticationTypes.USER,
refreshToken=st.session_state['refresh_token']
)
try:
response = await graphlit.client.query_share_point_folders(input, libraryId)
return response.share_point_folders.results, None
except GraphQLClientError as e:
return None, str(e)
Once we have our library and folder identifiers for the SharePoint folder we wish to ingest, we can create our SharePoint feed in Graphlit.
Here we are passing the refresh token and identifiers, and creating a recurrent feed which will check for new files in our folder, every 1 minute. This is done by specifying the schedulePolicy.
Also, we are assigning a content workflow, which provides instructions on how to prepare and extract the ingested content, as well as enriching the extracted entities.
async def create_feed(account_name, library_id, folder_id):
graphlit: Optional[Graphlit] = st.session_state['graphlit']
input = FeedInput(
name=f"{account_name}: {library_id}",
type=FeedTypes.SITE,
site=SiteFeedPropertiesInput(
type=FeedServiceTypes.SHARE_POINT,
sharePoint=SharePointFeedPropertiesInput(
authenticationType=SharePointAuthenticationTypes.USER,
refreshToken=st.session_state['refresh_token'],
accountName=account_name,
libraryId=library_id,
folderId=folder_id
)
),
workflow=EntityReferenceInput(
id=st.session_state['workflow_id']
),
schedulePolicy=FeedSchedulePolicyInput(
recurrenceType=TimedPolicyRecurrenceTypes.REPEAT,
repeatInterval="PT1M"
)
)
try:
response = await graphlit.client.create_feed(input)
st.session_state['feed_id'] = response.create_feed.id
return None
except GraphQLClientError as e:
return str(e)
GraphRAG Conversation
Before creating a conversation with an LLM, we have to create an LLM specification. Like with workflows, these are typically created once per application and reused across conversations.
Here we are saying to use the OpenAI GPT-4o model, with the EXTRACT_ENTITIES_GRAPH graph strategy, SECTION retrieval, and COHERE reranking.
EXTRACT_ENTITIES_GRAPH means to include the related entities (and their metadata) with the RAG context, vs EXTRACT_ENTITIES_FILTER which only uses the graph for filtering. SECTION retrieval means to expand any vector-related chunks into the surrounding section (i.e. page or semantic chunk). COHERE reranking will sort the content sources based on the user prompt, and filter out any irrelevant content sources, which may have inadvertently been seen as related by the vector search.
async def create_specification():
input = SpecificationInput(
name="Specification",
type=SpecificationTypes.COMPLETION,
serviceType=ModelServiceTypes.OPEN_AI,
searchType=SearchTypes.VECTOR,
openAI=OpenAIModelPropertiesInput(
model=OpenAIModels.GPT4O_128K,
temperature=0.1,
probability=0.2,
completionTokenLimit=2048,
),
graphStrategy=GraphStrategyInput(
type=GraphStrategyTypes.EXTRACT_ENTITIES_GRAPH
),
retrievalStrategy=RetrievalStrategyInput(
type=RetrievalStrategyTypes.SECTION
),
rerankingStrategy=RerankingStrategyInput(
serviceType=RerankingModelServiceTypes.COHERE
)
)
graphlit: Optional[Graphlit] = st.session_state['graphlit']
try:
response = await graphlit.client.create_specification(input)
st.session_state['specification_id'] = response.create_specification.id
except GraphQLClientError as e:
return str(e)
return None
We can now create our conversation, using this specification. You create a conversation once, and then prompt it multiple times, as the end-user provides their prompt. The user prompts and LLM completions are added as messages to the conversation, so you can use the entire conversation in your application user interface.
async def create_conversation():
input = ConversationInput(
name="Conversation",
specification=EntityReferenceInput(
id=st.session_state['specification_id']
)
)
graphlit: Optional[Graphlit] = st.session_state['graphlit']
try:
response = await graphlit.client.create_conversation(input)
st.session_state['conversation_id'] = response.create_conversation.id
except GraphQLClientError as e:
return str(e)
return None
Finally, we can prompt our conversation. Note that the knowledge graph related to the prompted conversation is returned along with the LLM response message.
async def prompt_conversation(prompt):
graphlit: Optional[Graphlit] = st.session_state['graphlit']
try:
response = await graphlit.client.prompt_conversation(prompt, st.session_state['conversation_id'])
message = response.prompt_conversation.message.message
graph = response.prompt_conversation.graph
return message, graph, None
except GraphQLClientError as e:
return None, None, str(e)
Graph Visualization
Now that we have our conversational knowledge graph, we can use PyVis in Streamlit to render it for viewing. We're using some helper functions to format the label, title, and assign icons to the node types.
def create_pyvis_conversation_graph(graph: Optional[PromptConversationPromptConversationGraph]):
g = create_pyvis_network()
for node in graph.nodes:
content_type = None
file_type = None
label = None
title = None
if node.type == EntityTypes.CONTENT:
content_type, file_type = parse_metadata(node.metadata)
label = parse_label(node.metadata)
title = parse_title(node.metadata)
shape = lookup_node_shape(node.type.name, content_type, file_type)
g.add_node(node.id, label=label if label is not None else node.name, shape=shape["shape"], icon=shape["icon"], color=lookup_node_color(node.type.name), title=title if title is not None else f'{node.type.name} [{node.id}]')
for edge in graph.edges:
# ensure start and end vertex exist in graph
if not edge.from_ in g.node_ids:
g.add_node(edge.from_)
if not edge.to in g.node_ids:
g.add_node(edge.to)
relation = format_relation(edge.relation)
width = 3 if edge.relation != "observed-by" else 1
g.add_edge(edge.from_, edge.to, label=relation, title=relation, width=width, arrowStrikethrough=False, arrows="middle")
return g
This lets us see the related content to our LLM conversation, and this graph gets updated every time the user provides a new prompt.
Wrapping Up
We have shown how to build a knowledge graph from content located on SharePoint, and how to have a GraphRAG conversation with the content and visualize the resulting knowledge graph.
We will be writing more articles on GraphRAG, including providing a more formal analysis on the quality of RAG vs GraphRAG responses, for various use cases.
Please let us know about your requirements for knowledge graphs, LLMs, and GraphRAG, and we'll be happy to learn more.
Summary
Please email any questions on this article or the Graphlit Platform to questions@graphlit.com.
For more information, you can read our Graphlit Documentation, visit our marketing site, or join our Discord community.
